Computational electrophysiology
2D simulations of cardiac tissue were started by coupling the cell models using gap junctional conductance for atria and ventricles. A matrix size of 100by100 using biophysical models takes approximately 8 hrs for one second simulation of cardiac activity. For reproducing ECG waveform from the 2D network an optimum path of conduction in atria and ventricles has to obtained. To construct a simplified representation of the cardiac conduction system simulations were done with reduced cell models.
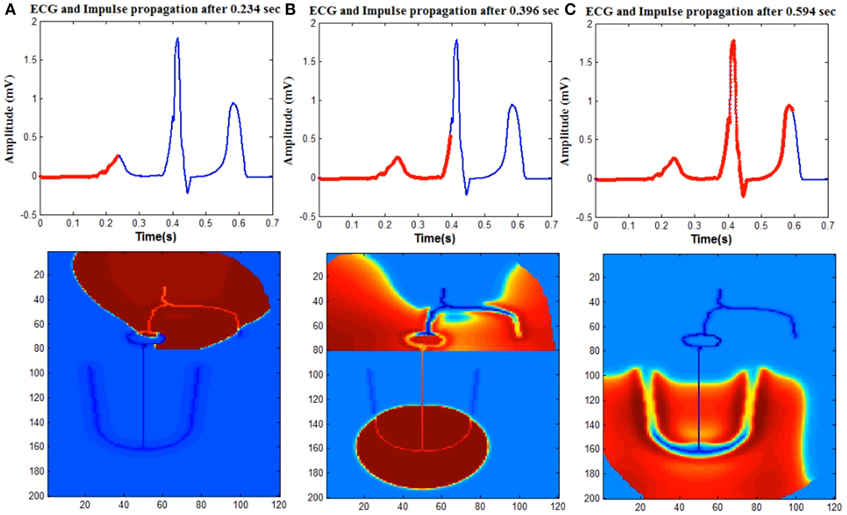
A simple 2D heart model using reduced cardiac cell models to reproduce cardiac electrical activity is proposed. The 2D structure contains anatomical and physiological details of the heart including various types of cardiac cells and the propagation pattern of the normal human heart. The components of the electrical conduction system of the heart that are modeled include SA (Sino atrial) node cells, fast conducting inter-atrial pathways, slow conducting AV(atrio-ventricular) node, Bundle of His cells, Purkinje network, atrial and ventricular myocardial cells.
SA nodal cells, Bundle of His cells and Purkinje cells are represented by the Fitzhugh Nagumo (FN) model which is a reduced model of the Hodgkin Huxley neuron model. For Bundle of His cells, the FN model is operated in excitable mode and for SA nodal cells and Purkinje cells it is modeled in oscillatory mode with frequency of 70 and 40 beats per minute respectively.
The atrial and ventricular myocardial cells are modeled by the Aliev- Panfilov (AP) two variable model proposed for cardiac excitation. The gap junction conductance is distributed optimally in the 2D geometry of the heart to produce a normal ECG signal. Arrhythmia conditions based on the conduction abnormalities also to be explained by the model by varying the gap junction conduction values.Finally the mapping the intrinsic conduction system of the heart has to be done using biophysical cell models. Human electrophysiological single cell models are available for various types of cardiac cells like atrial myocyte, ventricular myocyte, and purkinje fibers. The cells are coupled with gap junctions of varying conductance to produce a heterogeneous 2D network of cardiac cells to reproduce the electrical activity of the heart.
Simulation studies of cardiac arrhythmias at the whole heart level with electrocardiogram (ECG) gives an understanding of how the underlying cell and tissue level changes manifest as rhythm disturbances in the ECG. We present a 2D whole heart model (WHM2D) which can accommodate variations at the cellular level and can generate the ECG waveform. It is shown that, by varying cellular-level parameters like the gap junction conductance (GJC), excitability, action potential duration (APD) and frequency of oscillations of the auto-rhythmic cell in WHM2D a large variety of cardiac arrhythmias can be generated including sinus tachycardia, sinus bradycardia, sinus arrhythmia, sinus pause, junctional rhythm, Wolf Parkinson White syndrome and all types of AV conduction blocks. WHM2D includes key components of the electrical conduction system of the heart like the SA (Sino atrial) node cells, fast conducting intranodal pathways, slow conducting atriovenctricular (AV) node, bundle of His cells, Purkinje network, atrial, and ventricular myocardial cells. SA nodal cells, AV nodal cells, bundle of His cells, and Purkinje cells are represented by the Fitzhugh-Nagumo (FN) model which is a reduced model of the Hodgkin-Huxley neuron model. The atrial and ventricular myocardial cells are modeled by the Aliev-Panfilov (AP) two-variable model proposed for cardiac excitation. WHM2D can prove to be a valuable clinical tool for understanding cardiac arrhythmias.
Balakrishnan Minimol, Chakravarthy Srinivasa, Guhathakurta Soma "Simulation of cardiac arrhythmias using a 2D heterogeneous whole heart model" , Frontiers in Physiology, vol. 6, p. 00374 , 2015